Update: We congratulate the authors around Nathan Riguet (Lashuel lab, EPFL) for publishing their results in Nature Communications: https://www.nature.com/articles/s41467-021-26684-z
We were happy to help them with our 3DEMtrace pipeline: https://ariadne.ai/3demtrace/
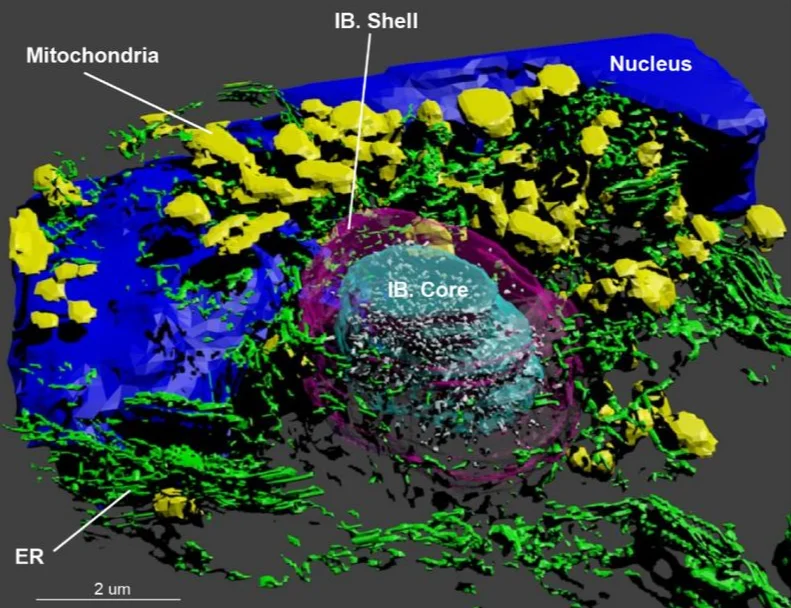
The pathogenesis of Huntington’s disease is accompanied by the formation of aggregates of the Huntingtin protein (Htt). This ground breaking study revealed that the aggregate formation has complex and far reaching consequences on the cellular machinery. Among other effects, it results in the recruitment and accumulation of dysfunctional membranous organelles. To investigate and compare the ultrastructural changes induced by Htt aggregates the authors used our 3dEMtrace pipeline for the automated segmentation of the cellular ultrastructure. More information available at: https://www.biorxiv.org/content/10.1101/2020.07.29.226977v1